Research
RNA-binding proteins involved in sRNA-dependent gene expression regulation in bacteria
The main interests of our laboratory are focused on RNA-binding proteins, which contribute to gene expression regulation by small regulatory RNAs (sRNAs) in bacteria. The aim of our studies is to explain how these small RNAs are recognized by specialized RNA-binding proteins, and what are the biologically important outcomes of these RNA-protein interactions.
Small RNAs are involved in bacterial adaptation to changing environmental conditions, maintenance of cellular homeostasis, and control of bacterial virulence. sRNAs are typically around 100 nucleotides long, and they control translation by binding complementary sequences within mRNAs. The action of sRNAs is dependent on RNA-binding proteins, which control sRNA stability, rearrange their structures, and facilitate their binding to mRNAs. In Gram-negative bacteria, such proteins include the Hfq chaperone, and the FinO-domain proteins (Figure 1), and in Gram-positive bacteria the KH-domain proteins.
The chaperone protein Hfq forms a homohexameric ring. The Hfq ring contains three distinct RNA binding sites that allow simultaneous binding of different RNAs. Depending on the mode of interactions and the identity of bound RNAs Hfq may either protect RNAs from degradation or accelerate their decay. Hfq can also serve as an RNA chaperone and facilitate the pairing between complementary sequences of two RNA molecules. In our studies we elucidated what properties of sRNA molecules make them more efficient competitors in binding to Hfq [1, 2], and also how Hfq facilitates the interactions between sRNA molecules and mRNA molecules [3, 4], or between sRNAs and anti-sRNAs [5].
The FinO domain proteins are present in Gram-negative bacteria, which also have Hfq [6]. These proteins consist of the FinO domain, which is named after the F-like plasmid FinO protein. Besides the FinO domain the proteins from this family may also contain additional N- or C-terminal extensions. The FinO domain contains the RNA binding site which recognizes the structures of transcription terminators. In our studies we described how the FinO domain of the E. coli ProQ protein recognizes RNA molecules, and proposed what RNA sequence elements prevent their binding by Hfq [7].
Much less is known about those RNA-binding proteins, which contribute to RNA-dependent regulation in Gram-positive bacteria. Recently, it has been proposed that such a role may be served by KH-domain proteins, named KhpA and KhpB, which are present together in numerous species of Gram-positive bacteria. These proteins bind numerous RNA molecules and participate in the regulation of bacterial cell division. However, the exact mode of their participation in the RNA-dependent gene expression regulation is not yet known [8].
A
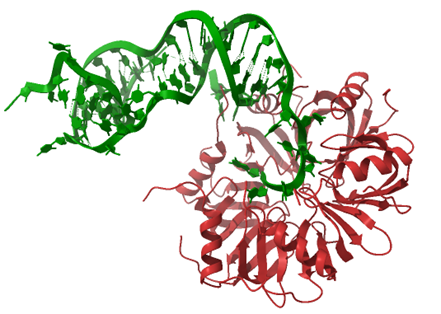
B

Figure 1. The interactions between RNA molecules and the chaperone protein Hfq (A) or the FinO domain of the RocC protein (B). (A) The structure of the complex of RydC sRNA (green) with the Hfq protein (red) from Salmonella (PDB: 4V2S). (B) The structure of the complex of RocR sRNA (green) with the FinO-domain of the RocC protein (orange) from Legionella (PDB: 7RGU).
References
- Olejniczak M. “Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance.” Biochemistry 50, 4427-4440 (2011)
- Małecka E.M., Stróżecka J., Sobańska D. and Olejniczak M. “Structure of bacterial regulatory RNAs determines their performance in competition for the chaperone protein Hfq.” Biochemistry 54, 1157-70 (2015)
- Wróblewska Z, Olejniczak M. “Hfq assists small RNAs in binding to the coding sequence of ompD mRNA and in rearranging its structure”, RNA (2016), 22(7):979-94
- Kwiatkowska J, Wróblewska Z, Johnson K.A., Olejniczak M. “The binding of Class II sRNA MgrR to two different sites on matchmaker protein Hfq enables efficient competition for Hfq and annealing to regulated mRNAs”. RNA 24(12), 1761-1784 (2018)
- Małecka E., Sobańska D., Olejniczak M. “Bacterial chaperone protein Hfq facilitates the annealing of sponge RNAs to small regulatory RNAs”. Journal of Molecular Biology 433(23), 167291 (2021)
- Olejniczak M., Storz G. “ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers?” Molecular Microbiology 104(6), 905-915 (2017)
- Stein E.M., Kwiatkowska J., Basczok M.M., Gravel C.M., Berry K.E., Olejniczak M. Determinants of RNA recognition by the FinO domain of the Escherichia coli ProQ protein. Nucleic Acids Research 48 (13), 7502-7519 (2020)
- Olejniczak M., Jiang X, Basczok MM, Storz G. KH domain proteins: Another family of bacterial RNA matchmakers? Molecular Microbiology 117(1):10-19 (2022)
